J. Cell Biol. 2011;193(5):917–933.
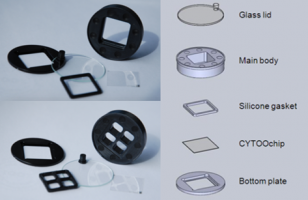
CYTOOchambers
CYTOOchambers 1-well:
The large imaging area of the 1-well gives access to over 10,000 cell patterns. This makes it ideal for when you have more the 4 different geometries printed on the chip (such as the CYTOOchip Starter’s or some custom chip layouts) and fo rare event measurements (like mitotic cells).
This chamber is also better suited for high resolution live cell imaging (with high aperture objectives)
CYTOOchambers 4-wells:
The 4-wells CYTOOchamber allows time-lapse or immunofluorescence experiments with multiple conditions in parallel while still providing thousands of patterns per well.
The 4-quandrants layout of CYTOOchips Arena and Motility make them fully compatible with the 4-well CYTOOchamber.
Advantages and key features
- Reusable and autoclavable
- Easy-to-use magnetic assembly
- 35 mm footprint
- Compatible with common microscope stages
- Up to 4 conditions in parallel
|
30-010 |
CYTOOchamber 1 well |
Ext. diameter 35 mm; with lid; compatible with CYTOOchip for Live Cell Imaging; 1 well; Max. imaging area 16 x 16 mm² |
|
30-011 |
CYTOOchamber 4 wells |
Ext. diameter 35 mm; with lid; compatible with CYTOOchip for Live Cell Imaging; 4 wells; Max. imaging area 7.5 x 7.5 mm² per well |
|
30-020 |
Microscope adaptor plate |
With clips; accepts up to 6 CYTOOchambers |
|
30-050 |
Silicone Gaskets for CYTOOchamber 1 well |
Compatible w/ CYTOOchamber Cat.No. 30-010, black, set of 2 |
|
30-051 |
Silicone Gaskets for CYTOOchamber 4 wells |
Compatible w/ CYTOOchamber Cat.No. 30-011, black, set of 2 |
Publications
Proc. Natl. Acad. Sci. U.S.A. 2012;109(5):1506–1511.
J Cell Biol. 2014 Apr 14;205(1):83-96. doi: 10.1083/jcb.201311104. Epub 2014 Apr 7.
Cell. 2013; Aug 15 [epub ahead of print].
J Biomol Screen. 2013; Aug 16 [epub ahead of print].
Biomaterials. 2013; Dec 04 [epub ahead of print].
Proc. Natl. Acad. Sci. U.S.A. 2006;103(52):19771–19776.
Dev. Cell. 2011;21(6):1171–1178.
Molecular Cell - 27 February 2014
Cancer Res. 2011;71(1):134–142.
Mol. Biol. Cell. 2012; Dec 05 [epub ahead of print].
J Cell Biol. 2012;198(6):1011-1023.
Cell Motil. Cytoskeleton. 2006;63(6):341–355
J. Cell Biol. 2010;191(2):303–312.
J. Cell Biol. 2010;191(2):233–236.
Methods in Cell Biology.Vol Volume 118. Methods for Analysis of Golgi Complex Function. Academic Press; 2013:105–123.
Methods in Cell Biology. Vol 115. Academic Press; 2013:97–108.
Nat. Cell Biol. 2012;14(3):311–317.
Cell. 2013;154(2):391–402.
J Cell Biol. 2012;199(1):97.
Dev Cell. 2012;23(6):1153–1166.
Nature. 2007;447(7143):493–496.
Plos One. 2012;7(8):e40864.
PLoS ONE. 2012;7(8):e40864.
ACS Nano, 2014, 8 (3), pp 2033–2047 Publication Date (Web): February 2, 2014
Cell Rep. 2013;Oct 17 [epub ahead of print].
J Vis Exp. 2010;(46):pii: 2514.
Pflugers Arch. 2011;462(1):75–87.
PLoS ONE. 2012;7(5):e36894.
Neoplasia. 2012;14(8):771–777.
FASEB J. 2012;26(6):2592–2606.
PLoS ONE. 2013;8(2):e56231.
Nat. Cell Biol. 2011;13(9):1040–1050.
Small GTPases. 2013;4(2).
J Mol Biol. 2011;405(4-3):1004–1026.
Curr Opin Cell Biol. 2013;Aug 16 [epub ahead of print].
Genes Dev. 2008;22(16):2189–2203.
Lab Chip. 2013;13(4):714–721.
Nat Commun. 2013;4(2252).
J. Cell. Sci. 2010;123(24):4201–4213.
FEBS J. 2013; June 27 [epub ahead of print].
J Clin Invest. 2013; Mar 15 [epub ahead of print].
J Biomater Tissue Eng. 2013;3(4):461–471
Microbes Infect. 2012; Nov 02 [epub ahead of print].
Nat. Methods. 2010;7(7):560–566.
PLoS ONE. 2012;7(9):e46265.
IntraVital. 2012;1(1):77-85.
J. Cell. Sci. 2012;125(9):2134–2140.
Nature. 2011;474(7350):179–183.
J. Cell. Sci. 2011;124(22):3884–3893.
J. Biochem. Cell Biol. 2012;44(6):980–988.
J Biol Chem. 2013;288(38):27619–27637.
Nat. Cell Biol. 2005;7(10):947–953.
J. Biol. Chem. 2011;286(30):26743–26753.
Hum. Mol. Genet. 2011;20(24):4840–4850
Nat Cell Biol. 2013; May 26 [epub ahead of print] .